Chapter 2 Getting Started with cisDynet

2.1 Why use cisDynet?
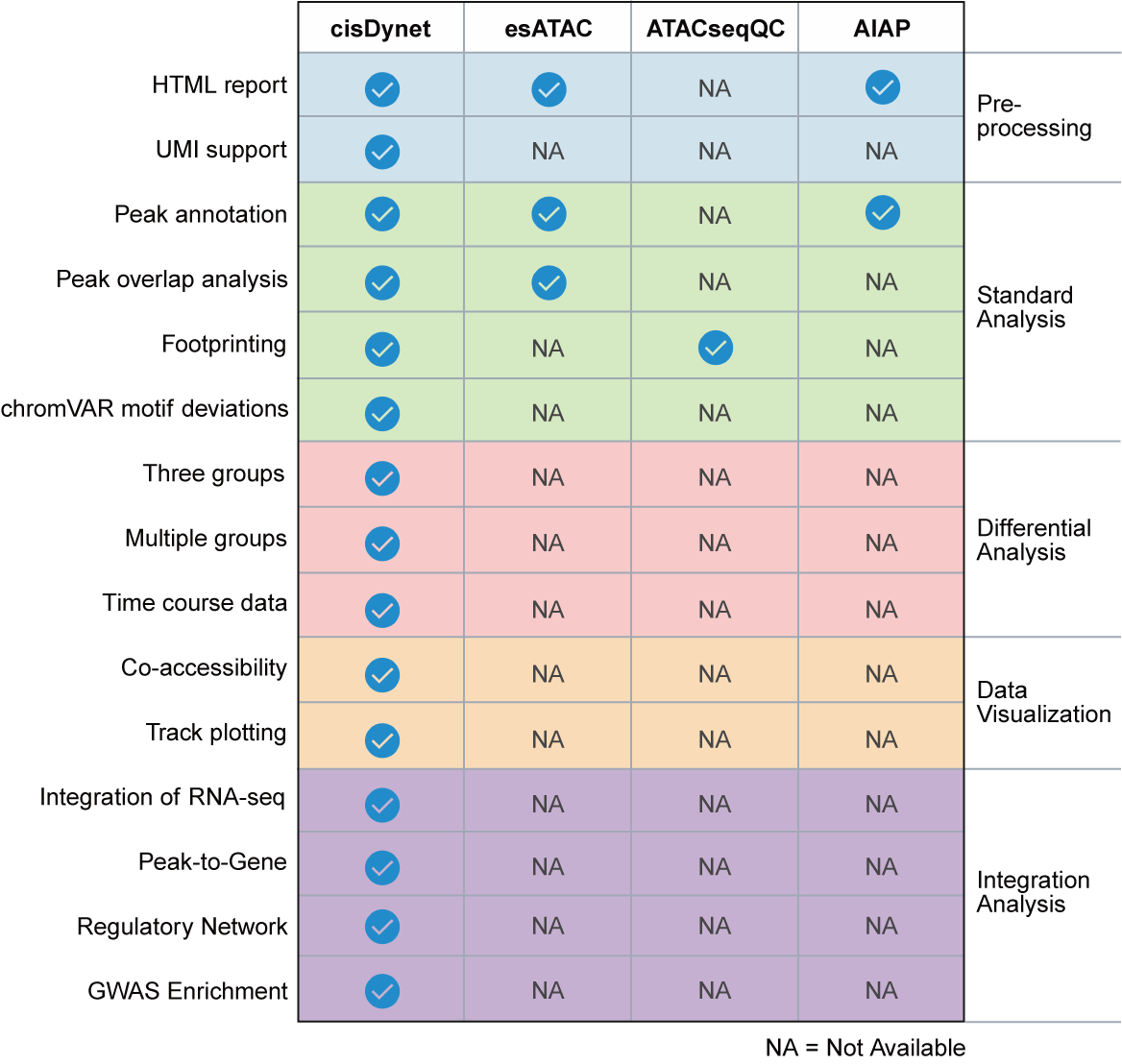
There are several ATAC-seq analysis tools available, so why use cisDynet? Currently these software focus more on QC/pre-processing of data, with little or no downstream analysis involved. Here, our cisDynet offers features not available in other tools, especially advanced downstream analysis. We also provide extensive data visualization capabilities.
Our whole workflow consists of two major parts, one is data preprocessing and QC (cisDynet_snakemake) built on snakemake framework, and the other is downstream analysis based on R package (cisDynet).
2.2 Getting Set Up
Before you use cisDynet, use the addAnnotation and addFootprint functions to add the necessary annotations.
library(cisDynet)##
##
## ▄▄▄▄▄▄▄▄▄▄▄ ▄▄▄▄▄▄▄▄▄▄▄ ▄▄▄▄▄▄▄▄▄▄▄ ▄▄▄▄▄▄▄▄▄▄ ▄ ▄ ▄▄ ▄ ▄▄▄▄▄▄▄▄▄▄▄ ▄▄▄▄▄▄▄▄▄▄▄
## ▐░░░░░░░░░░░▌▐░░░░░░░░░░░▌▐░░░░░░░░░░░▌▐░░░░░░░░░░▌ ▐░▌ ▐░▌▐░░▌ ▐░▌▐░░░░░░░░░░░▌▐░░░░░░░░░░░▌
## ▐░█▀▀▀▀▀▀▀▀▀ ▀▀▀▀█░█▀▀▀▀ ▐░█▀▀▀▀▀▀▀▀▀ ▐░█▀▀▀▀▀▀▀█░▌▐░▌ ▐░▌▐░▌░▌ ▐░▌▐░█▀▀▀▀▀▀▀▀▀ ▀▀▀▀█░█▀▀▀▀
## ▐░▌ ▐░▌ ▐░▌ ▐░▌ ▐░▌▐░▌ ▐░▌▐░▌▐░▌ ▐░▌▐░▌ ▐░▌
## ▐░▌ ▐░▌ ▐░█▄▄▄▄▄▄▄▄▄ ▐░▌ ▐░▌▐░█▄▄▄▄▄▄▄█░▌▐░▌ ▐░▌ ▐░▌▐░█▄▄▄▄▄▄▄▄▄ ▐░▌
## ▐░▌ ▐░▌ ▐░░░░░░░░░░░▌▐░▌ ▐░▌▐░░░░░░░░░░░▌▐░▌ ▐░▌ ▐░▌▐░░░░░░░░░░░▌ ▐░▌
## ▐░▌ ▐░▌ ▀▀▀▀▀▀▀▀▀█░▌▐░▌ ▐░▌ ▀▀▀▀█░█▀▀▀▀ ▐░▌ ▐░▌ ▐░▌▐░█▀▀▀▀▀▀▀▀▀ ▐░▌
## ▐░▌ ▐░▌ ▐░▌▐░▌ ▐░▌ ▐░▌ ▐░▌ ▐░▌▐░▌▐░▌ ▐░▌
## ▐░█▄▄▄▄▄▄▄▄▄ ▄▄▄▄█░█▄▄▄▄ ▄▄▄▄▄▄▄▄▄█░▌▐░█▄▄▄▄▄▄▄█░▌ ▐░▌ ▐░▌ ▐░▐░▌▐░█▄▄▄▄▄▄▄▄▄ ▐░▌
## ▐░░░░░░░░░░░▌▐░░░░░░░░░░░▌▐░░░░░░░░░░░▌▐░░░░░░░░░░▌ ▐░▌ ▐░▌ ▐░░▌▐░░░░░░░░░░░▌ ▐░▌
## ▀▀▀▀▀▀▀▀▀▀▀ ▀▀▀▀▀▀▀▀▀▀▀ ▀▀▀▀▀▀▀▀▀▀▀ ▀▀▀▀▀▀▀▀▀▀ ▀ ▀ ▀▀ ▀▀▀▀▀▀▀▀▀▀▀ ▀## Author: Tao Zhu
## cisDynet: Version 1.0.0
## If you encounter a bug please report : https://github.com/tzhu-bio/cisDynet/issuesaddAnnotation(gene_bed = "F:/cisDynet/example/hg19_gene_standard.bed",
gtf = "F:/cisDynet/data/bw/hg19.gtf",
genome_size = "F:/cisDynet/example/hg19.chrom.size")addFootprint(corrected_signal = "F:/cisDynet/example/ATACcorrect/",
bindetect_result = "F:/cisDynet/example/BINDetect/")Where the gene_bed is the gene’s bed file, including chromosome number, start position, end position, and so on. Note that column names are NOT included.
## V1 V2 V3 V4 V5 V6
## 1 chr1 11869 14412 ENSG00000223972 1 +
## 2 chr1 14363 29806 ENSG00000227232 1 -
## 3 chr1 52473 54936 ENSG00000268020 1 +
## 4 chr1 62948 63887 ENSG00000240361 1 +
## 5 chr1 69091 70008 ENSG00000186092 1 +
## 6 chr1 131025 134836 ENSG00000233750 1 +Where the genome_size file includes the size of each chromosome. This file has been generated by our snakemake and you can read it directly.
## V1 V2
## 1 chr1 249250621
## 2 chr2 243199373
## 3 chr3 198022430
## 4 chr4 191154276
## 5 chr5 180915260
## 6 chr6 171115067