Chapter 10 Motif Enrichment
In the above chapter we know which OCRs are differentiated by compared to the treatment group, or the dynamics of changing OCRs in different periods/samples. Now we’re more interested in knowing exactly what kind of motifs these changing OCRs will have. In other words, what exactly are the important TFs that determine the differences between your different samples?
The most common way to do this is to use PWM / PFM to go against the sequences in your differential/dynamic OCR to see which TF-bound motifs match the sequences in your differential OCRs. There are already many tools available to help us do this. Common ones are FIMO, AME and SEA in the MEME toolkit, to name a few. So we’re not developing a new approach here, what we’re doing is taking existing tools and making them work well and compatible with our cisDynet. Here, we will mainly consider AME, FIMO, TOMTOM from the MEME toolkit. thankfully, Nystrom SL has implemented these functions in R already. We will further wrap these functions to match our cisDynet.
10.1 ChromVAR
counts <- getCount(sample_list=c("Bulk_B",
"Mem_B",
"Naive_B",
"Plasmablasts",
"CD8pos_T",
"Central_memory_CD8pos_T",
"Effector_memory_CD8pos_T",
"Naive_CD8_T",
"Gamma_delta_T",
"Effector_CD4pos_T",
"Follicular_T_Helper",
"Memory_Teffs",
"Memory_Tregs",
"Naive_Teffs",
"Regulatory_T",
"Th1_precursors",
"Immature_NK",
"Mature_NK",
"Monocytes",
"pDCs"),
cut_path="F:/cisDynet/example/cut_sites/",
peak_path="F:/cisDynet/example/peaks",
save_file_path="F:/cisDynet/example/",
peak_suffix = "_peaks_unique.narrowPeak.bed")
head(counts)## Bulk_B Mem_B Naive_B Plasmablasts CD8pos_T
## chr1:9910-10596 119 152 103 185 163
## chr1:564574-570175 454341 410439 515101 280478 526103
## chr1:713757-714733 1496 1341 1621 991 1288
## chr1:750692-750842 2 0 0 0 0
## chr1:752562-752915 14 14 4 30 19
## chr1:762339-763292 490 477 564 381 480
## Central_memory_CD8pos_T Effector_memory_CD8pos_T Naive_CD8_T
## chr1:9910-10596 135 154 50
## chr1:564574-570175 266369 425560 396896
## chr1:713757-714733 1255 1284 451
## chr1:750692-750842 0 5 0
## chr1:752562-752915 41 53 6
## chr1:762339-763292 361 451 136
## Gamma_delta_T Effector_CD4pos_T Follicular_T_Helper
## chr1:9910-10596 116 126 144
## chr1:564574-570175 254157 495122 361234
## chr1:713757-714733 1182 1546 816
## chr1:750692-750842 4 0 0
## chr1:752562-752915 51 46 21
## chr1:762339-763292 551 465 260
## Memory_Teffs Memory_Tregs Naive_Teffs Regulatory_T
## chr1:9910-10596 129 237 74 154
## chr1:564574-570175 300415 295119 421663 347173
## chr1:713757-714733 1235 569 885 397
## chr1:750692-750842 1 0 0 0
## chr1:752562-752915 72 14 7 6
## chr1:762339-763292 463 207 290 163
## Th1_precursors Immature_NK Mature_NK Monocytes pDCs
## chr1:9910-10596 114 91 128 160 53
## chr1:564574-570175 400776 104315 138450 172779 96076
## chr1:713757-714733 1089 886 836 461 112
## chr1:750692-750842 0 13 3 0 0
## chr1:752562-752915 18 49 24 46 10
## chr1:762339-763292 392 651 558 164 31fragment_counts <- makeSummarizedExperiment(counts)## 2023-10-23 15:31:51 Making SummarizedExperiment object...library(chromVAR)
library(motifmatchr)
library(BSgenome.Hsapiens.UCSC.hg19)fragment_counts <- addGCBias(fragment_counts, genome = BSgenome.Hsapiens.UCSC.hg19)motifs <- getJasparMotifs2018()motif_ix <- matchMotifs(motifs, fragment_counts, genome = BSgenome.Hsapiens.UCSC.hg19)dev <- computeDeviations(object = fragment_counts, annotations = motif_ix)dev_score <- deviationScores(dev)pheatmap::pheatmap(dev_score[1:20,])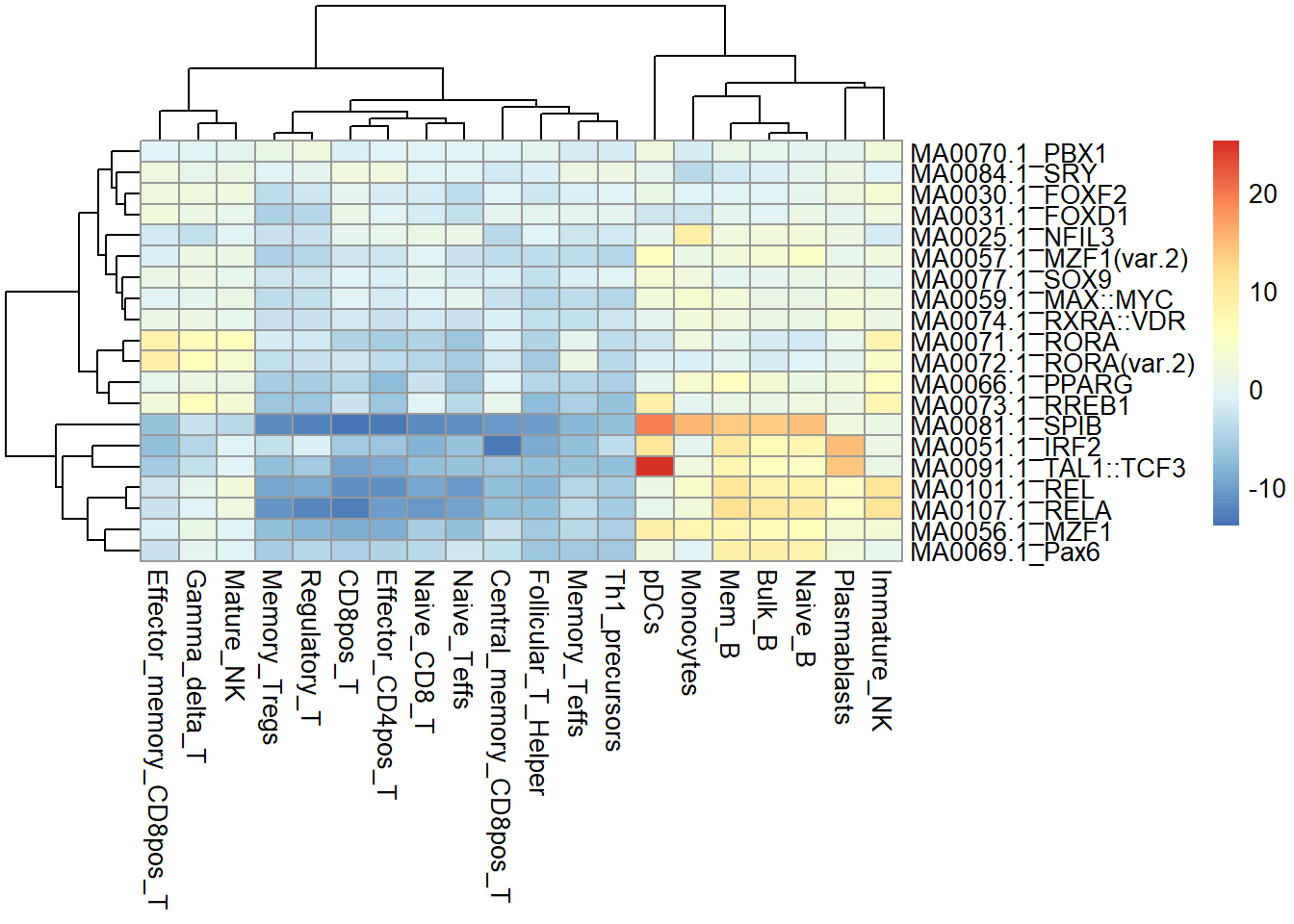
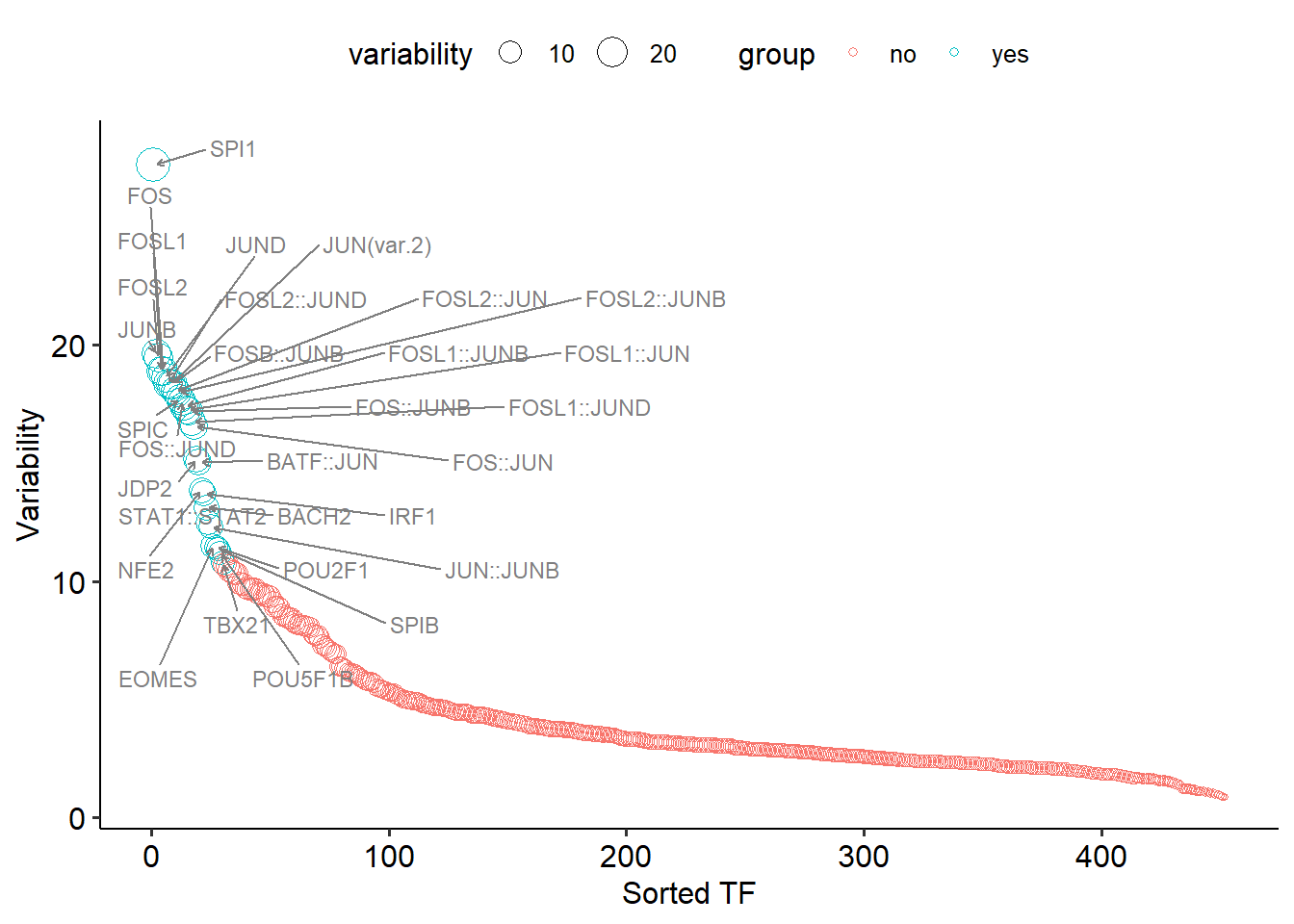
getRankMotif(deviation_result = dev)## 2023-10-23 15:35:37 Computing the variability...