cisDynet:an integrated platform for modeling gene-regulatory dynamics and networks
2023-08-24
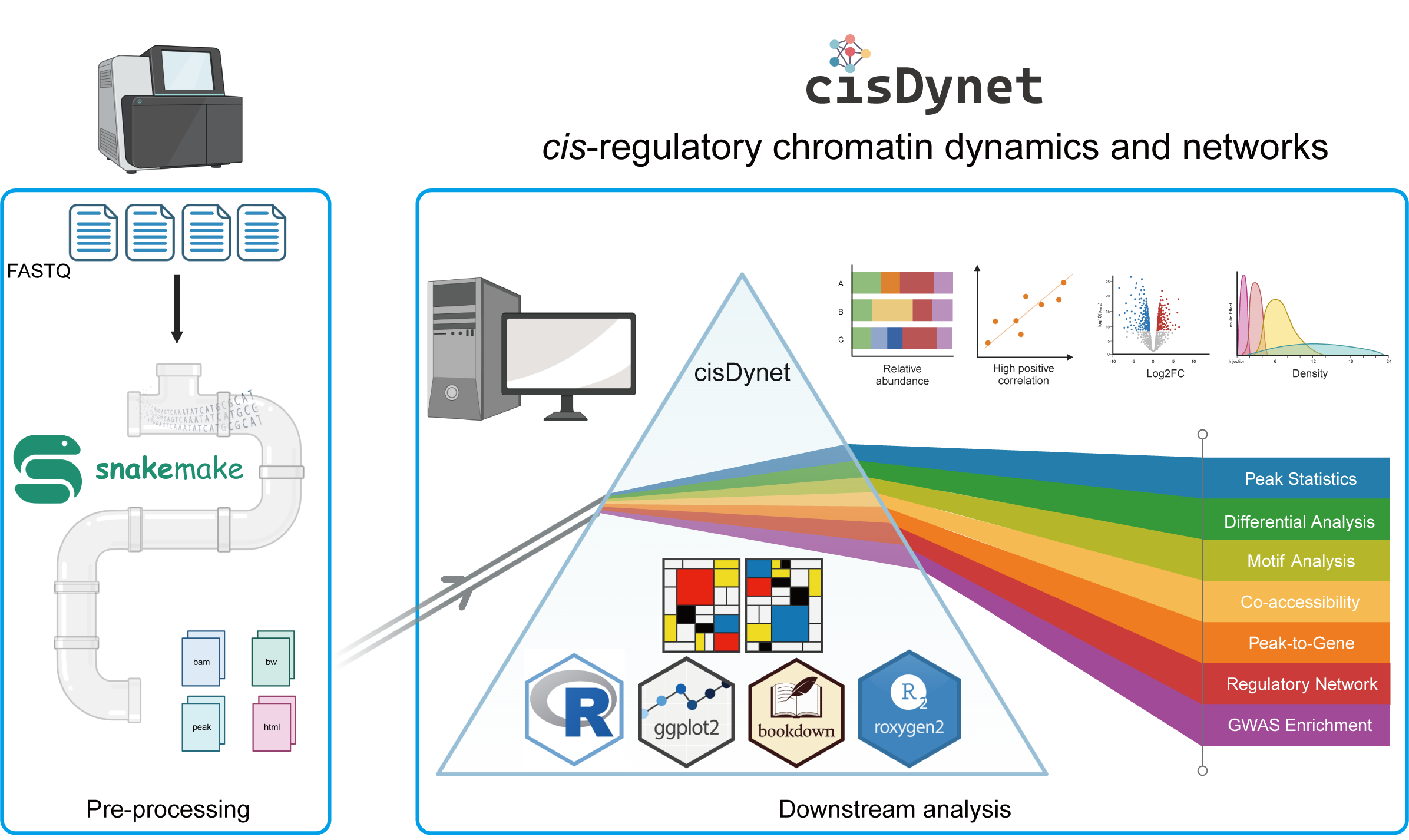
Chapter 1 Primer
Chromatin accessibility sequencing is a powerful technique that allows us to identify open regions within the genome, providing valuable insights into regulatory elements and enabling the construction of intricate regulatory networks. Although several tools are available for processing chromatin accessibility data, most of them primarily focus on data pre-processing and rarely involve downstream analyses (such as PEPATAC, esATAC, etc.). To bridge this gap, we have introduced cisDynet, a comprehensive solution that combines efficient data pre-processing through Snakemake and R functions with advanced downstream analysis capabilities. cisDynet excels in conventional data analyses, encompassing peak statistics, peak annotation, differential analysis, motif enrichment analysis and more. Additionally, it allows to perform sophisticated data exploration such as tissue-specific peak identification, time-course data fitting, integration of RNA-seq data to establish peak-to-gene associations, constructing regulatory networks, and conducting enrichment analysis of GWAS variants.